Cells, Free Full-Text
Por um escritor misterioso
Descrição
Identifying tissue-specific molecular signatures of active regulatory elements is critical to understanding gene regulatory mechanisms. In this study, transcription start sites (TSS) and enhancers were identified using Cap analysis of gene expression (CAGE) across endometrial stromal cell (ESC) samples obtained from women with (n = 4) and without endometriosis (n = 4). ESC TSSs and enhancers were compared to those reported in other tissue and cell types in FANTOM5 and were integrated with RNA-seq and ATAC-seq data from the same samples for regulatory activity and network analyses. CAGE tag count differences between women with and without endometriosis were statistically tested and tags within close proximity to genetic variants associated with endometriosis risk were identified. Over 90% of tag clusters mapping to promoters were observed in cells and tissues in FANTOM5. However, some potential cell-type-specific promoters and enhancers were also observed. Regions of open chromatin identified using ATAC-seq provided further evidence of the active transcriptional regions identified by CAGE. Despite the small sample number, there was evidence of differences associated with endometriosis at 210 consensus clusters, including IGFBP5, CALD1 and OXTR. ESC TSSs were also located within loci associated with endometriosis risk from genome-wide association studies. This study provides novel evidence of transcriptional differences in endometrial stromal cells associated with endometriosis and provides a valuable cell-type specific resource of active TSSs and enhancers in endometrial stromal cells.

Cell-free DNA tissues of origin by methylation profiling reveals significant cell, tissue, and organ-specific injury related to COVID-19 severity - ScienceDirect
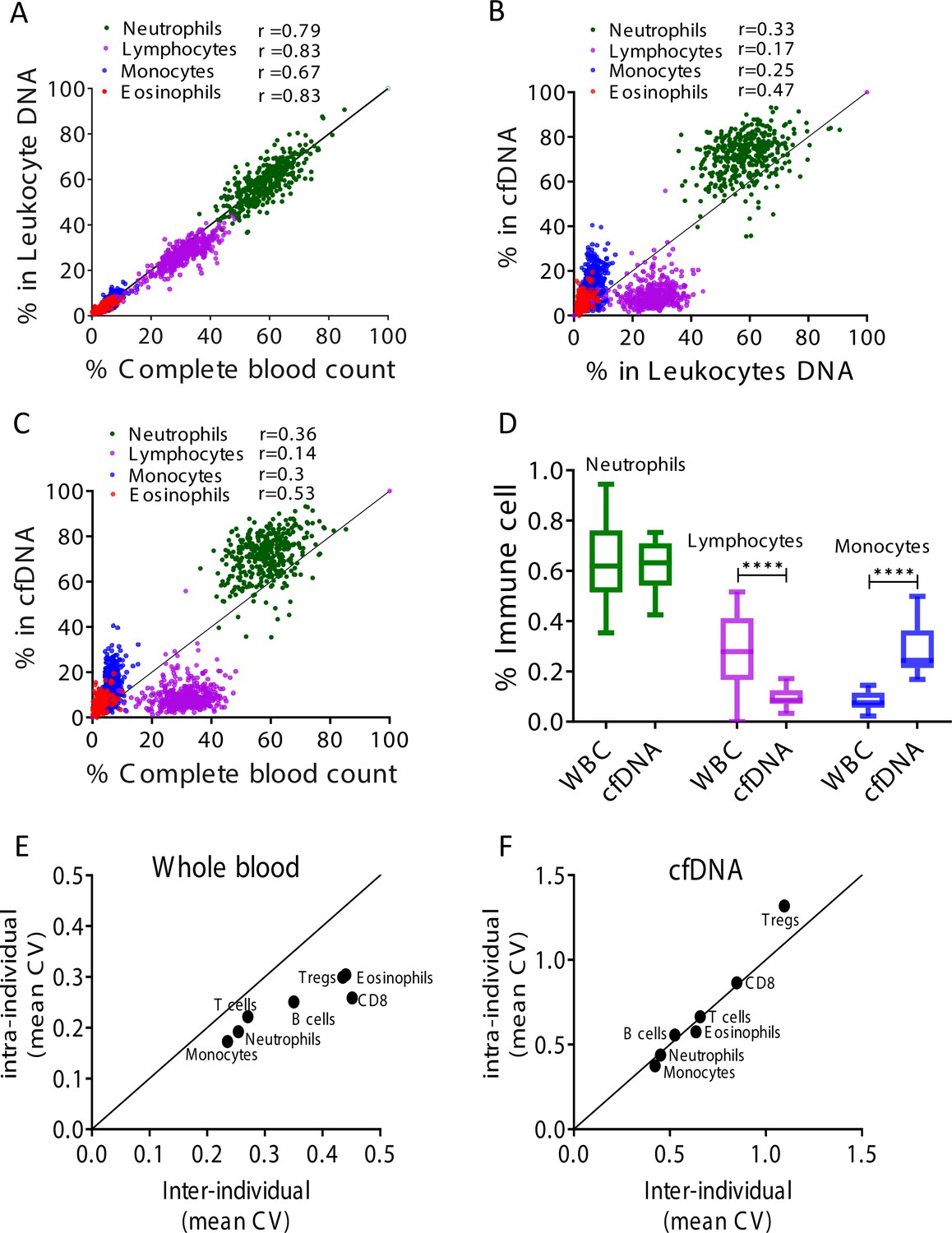
Remote immune processes revealed by immune-derived circulating cell-free DNA
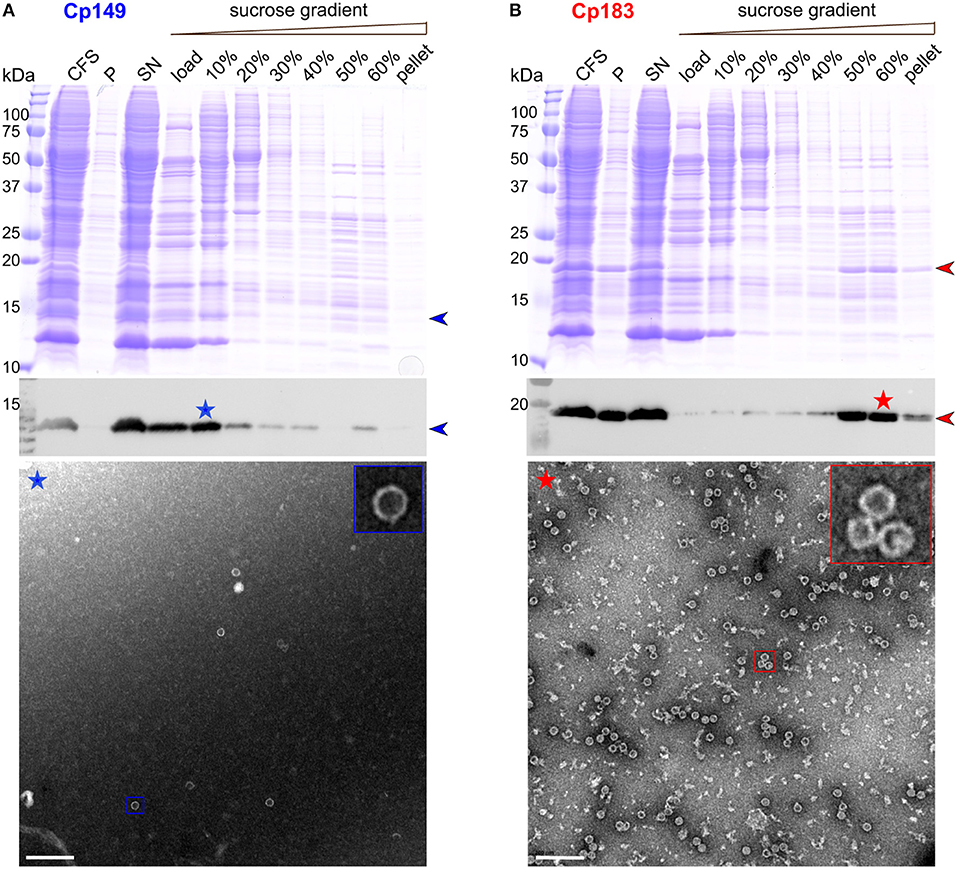
Frontiers Combining Cell-Free Protein Synthesis and NMR Into a Tool to Study Capsid Assembly Modulation
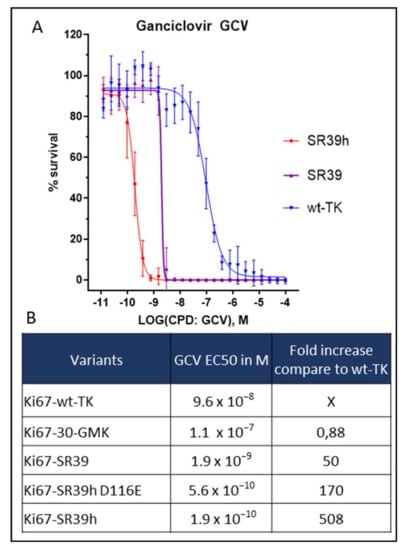
Serial Number Alcohol 120 1.9 6 - Colaboratory
Nature Reviews Molecular Cell Biology on X: From May cover Challenges and directions in studying cell–cell communication by #ExtracellularVesicles @DaveCarter1234 @guillaume_niel @VaderPieter Clayton Lambert & Raposo #EVsAreCool FREE pdf: https

Cell-free mutant analysis combined with structure prediction of a lasso peptide biosynthetic protein B2

Cell-Free Translation Systems
Cells, Free Full-Text

Label-free, full-field visualization of red blood cell (RBC)

Rapid cell-free characterization of multi-subunit CRISPR effectors and transposons - ScienceDirect
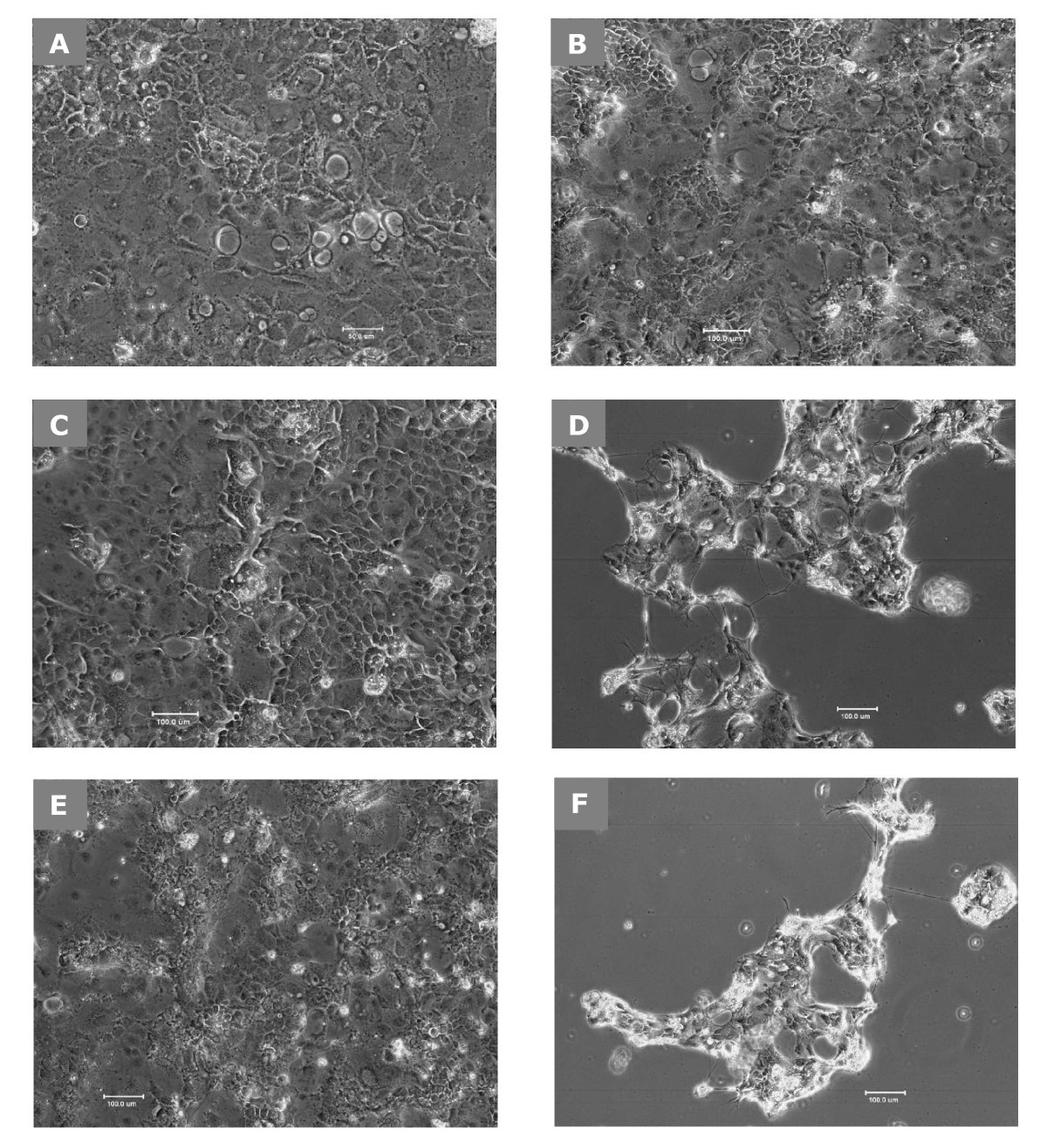
Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells, Gut Pathogens
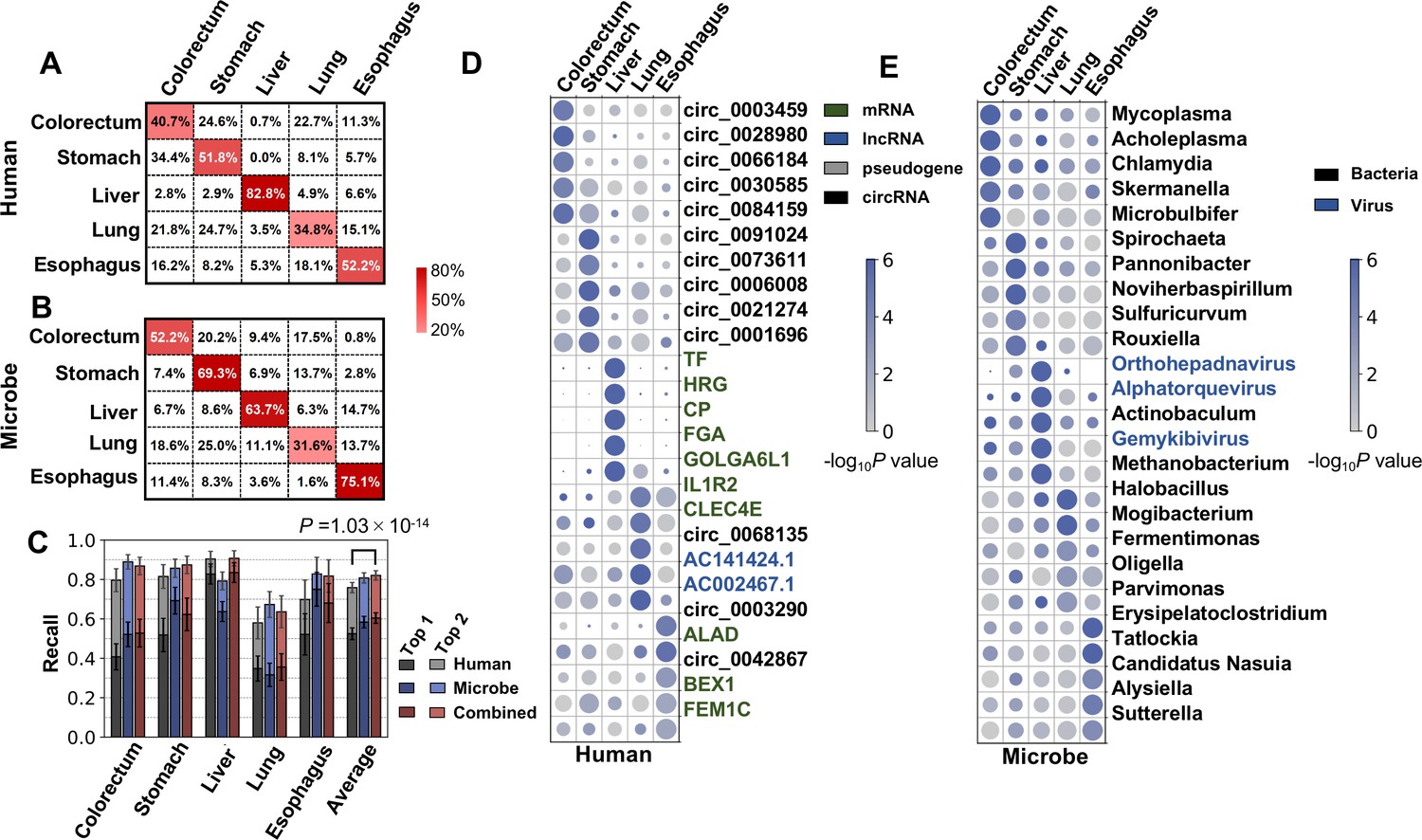
Cancer type classification using plasma cell-free RNAs derived from human and microbes
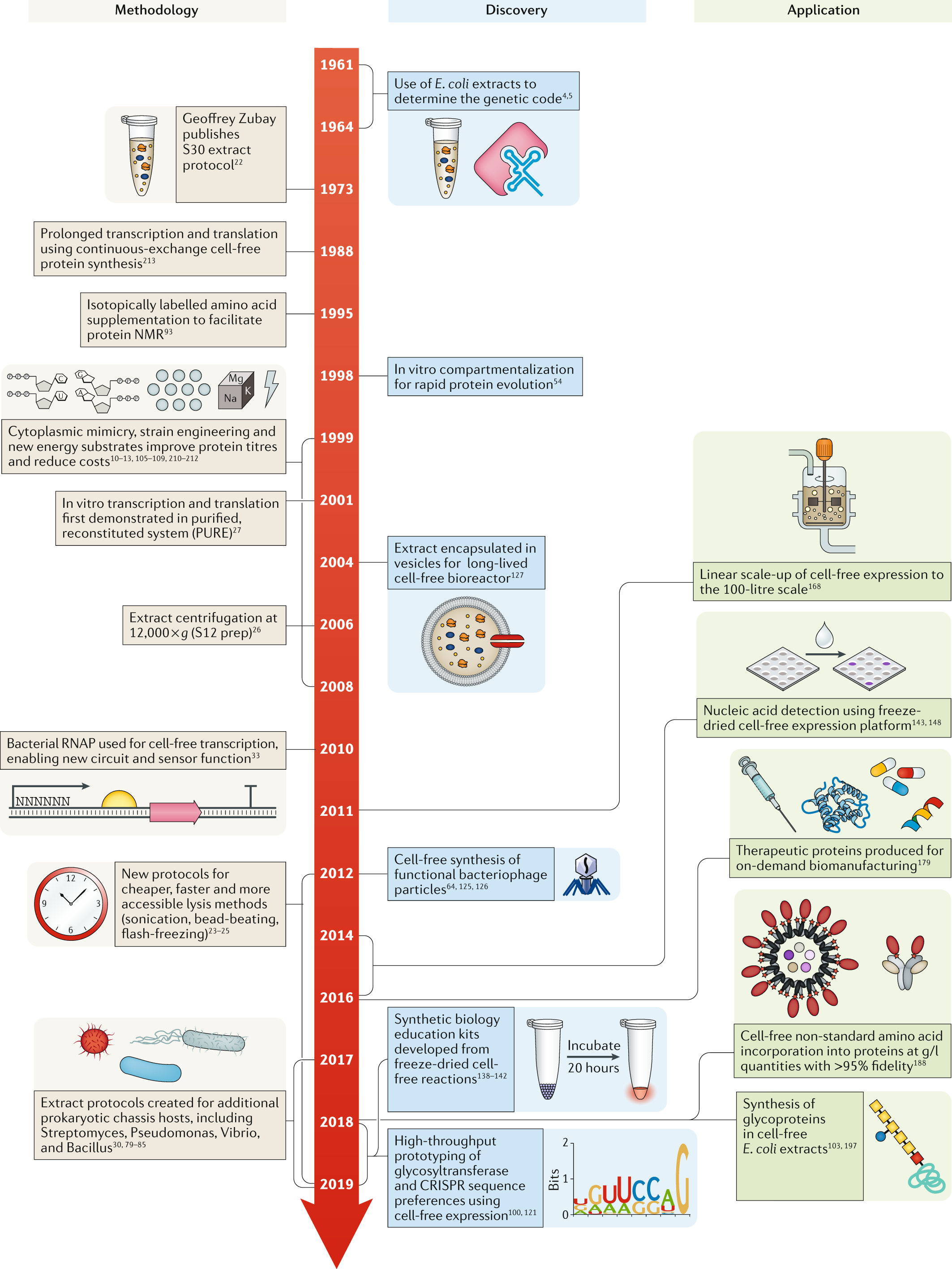
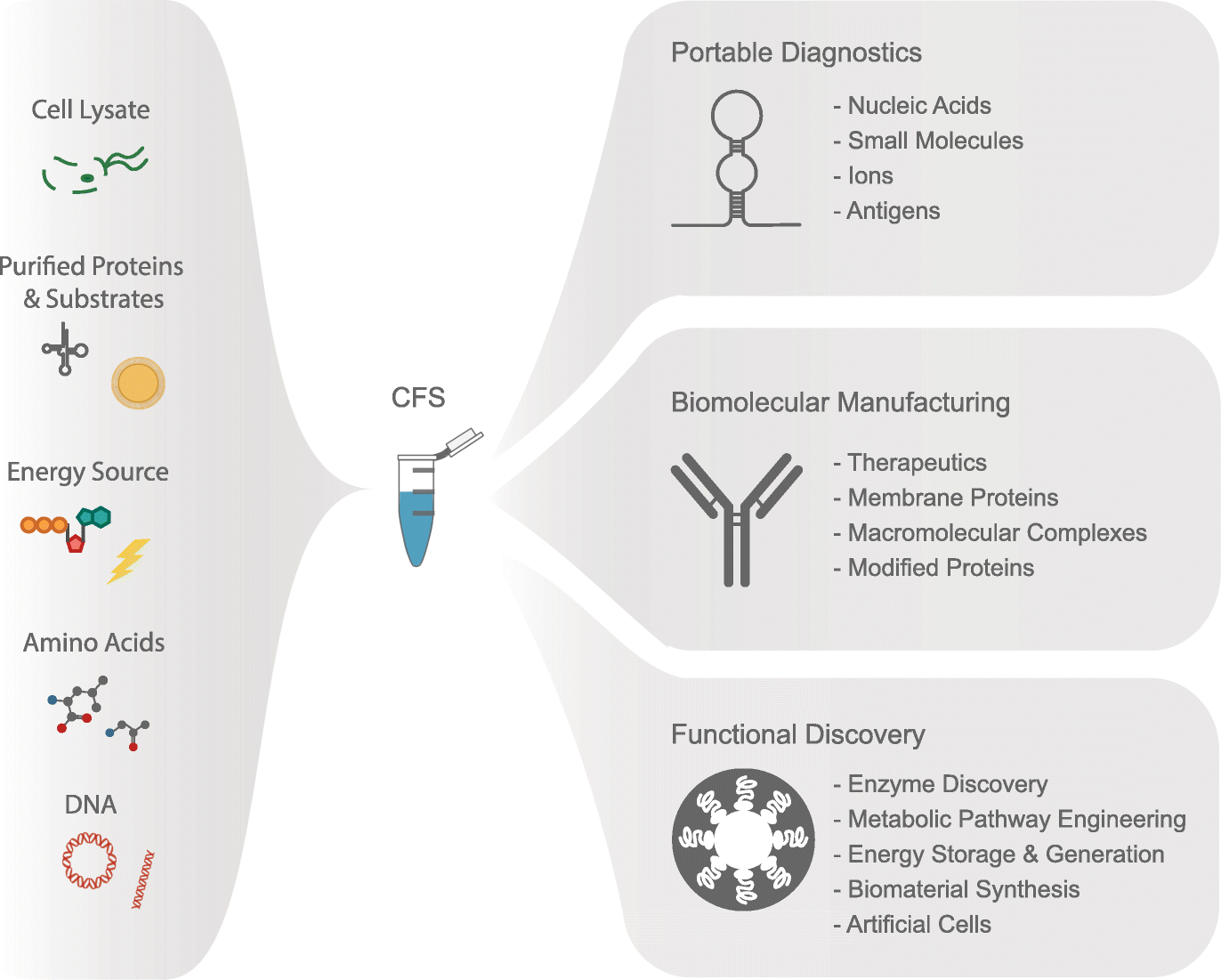
Cell-free gene expression: an expanded repertoire of applications
THE LIVES OF A CELL : LEWIS THOMAS : Free Download, Borrow, and Streaming : Internet Archive
from Flow Cytometry to Cytomics, Page 2
de
por adulto (o preço varia de acordo com o tamanho do grupo)




)


