Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Descrição
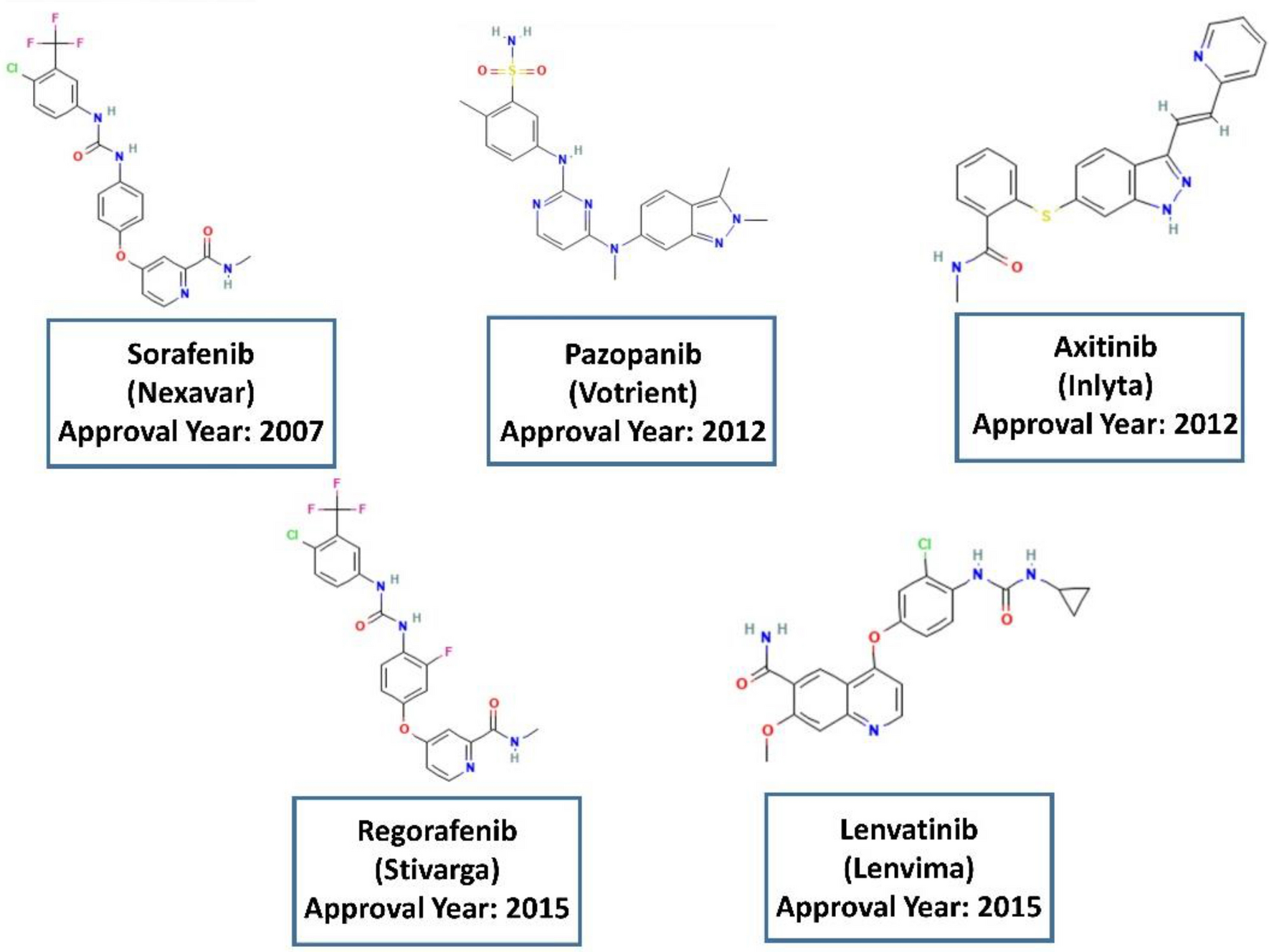
The use of machine learning modeling, virtual screening, molecular docking, and molecular dynamics simulations to identify potential VEGFR2 kinase inhibitors

Deep mutational scanning and machine learning reveal structural and molecular rules governing allosteric hotspots in homologous proteins
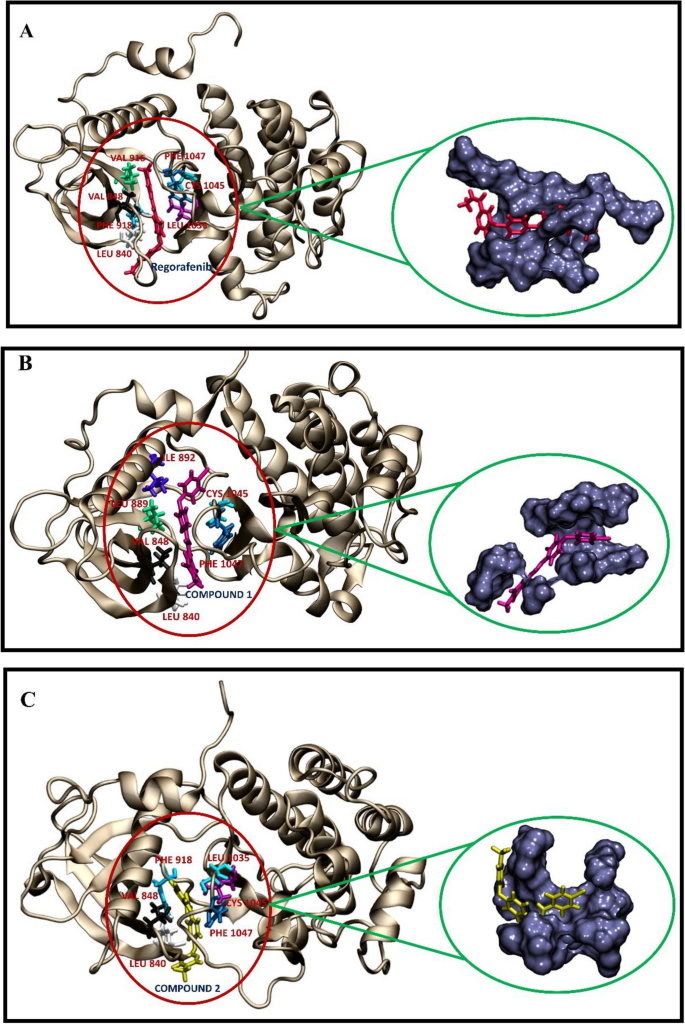
The use of machine learning modeling, virtual screening, molecular docking, and molecular dynamics simulations to identify potential VEGFR2 kinase inhibitors
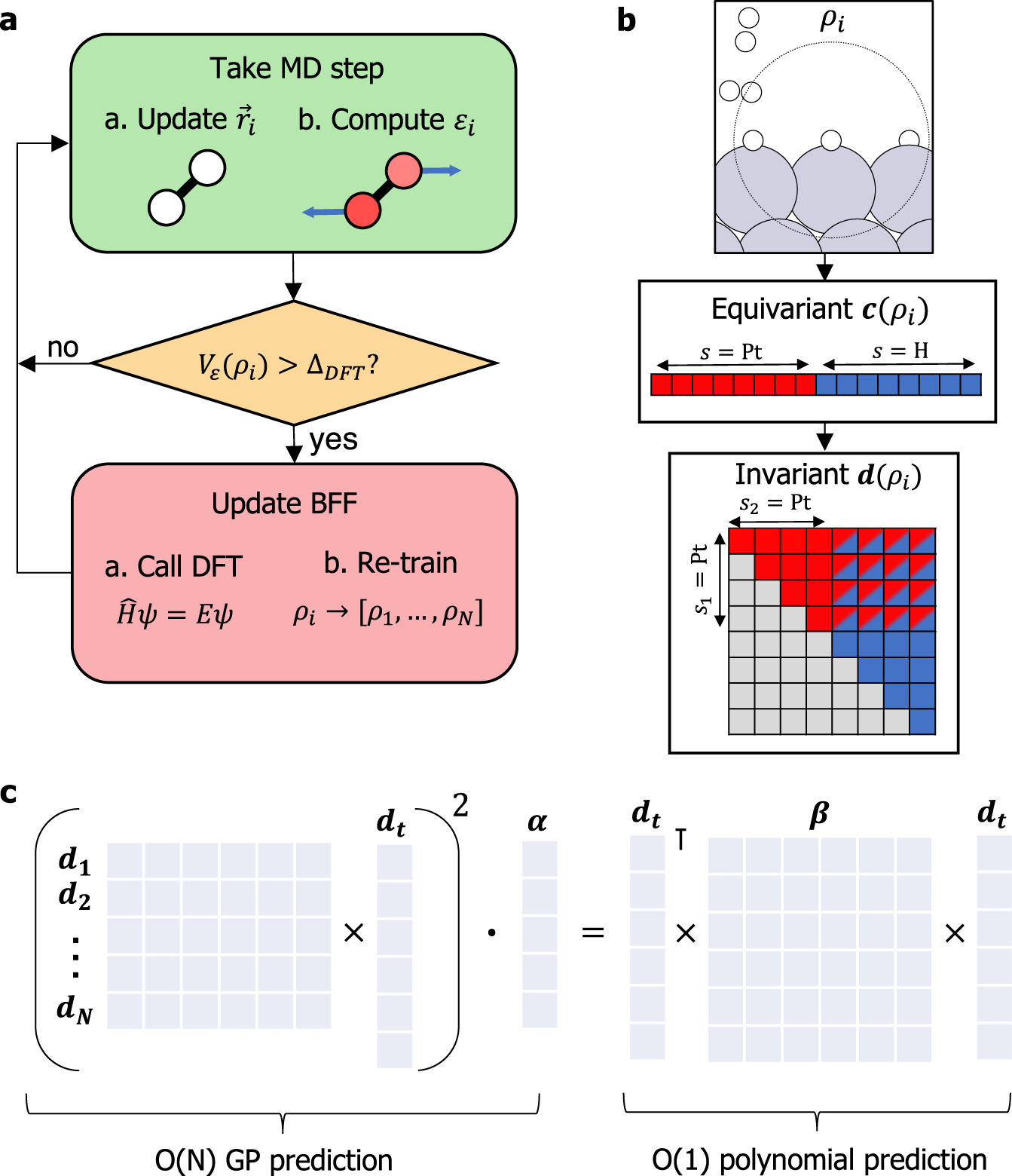
Active learning of reactive Bayesian force fields applied to heterogeneous catalysis dynamics of H/Pt

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Deep learning-based molecular dynamics simulation for structure-based drug design against SARS-CoV-2 - ScienceDirect

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
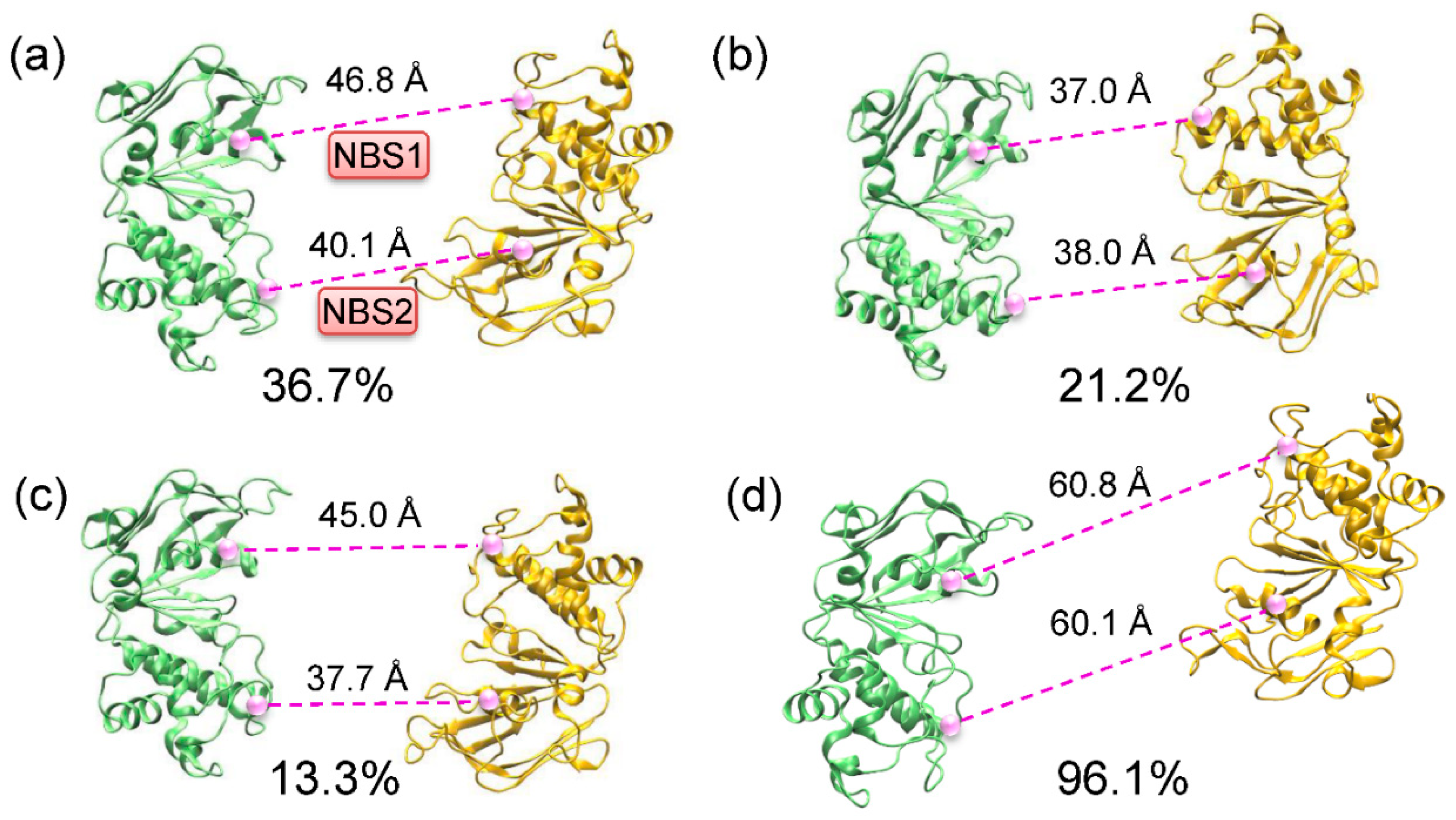
Molecules, Free Full-Text
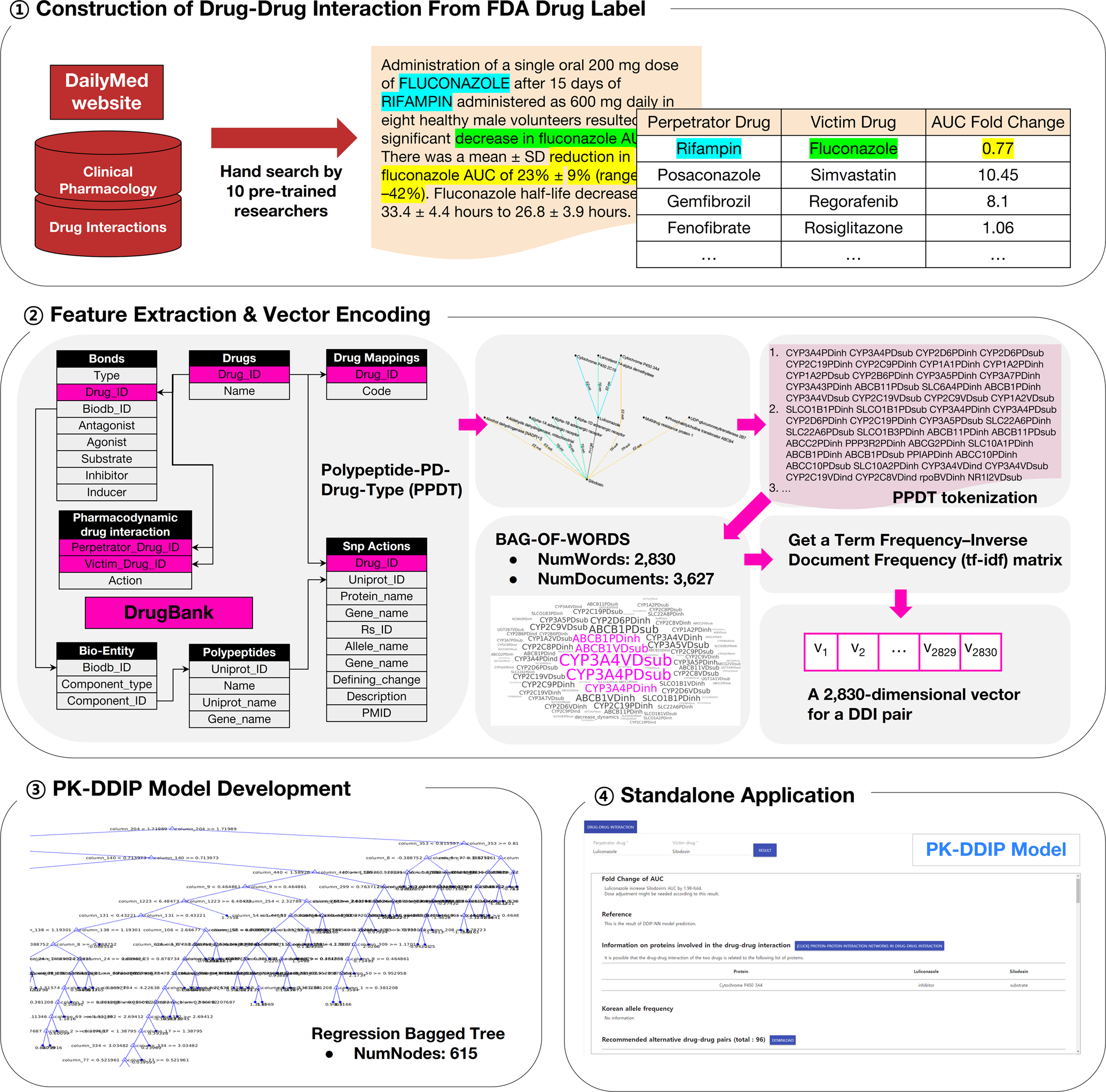
Machine learning-based quantitative prediction of drug exposure in drug-drug interactions using drug label information

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Plot of K p,brain and K p,uu,brain in the P-gp substrate before and

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
de
por adulto (o preço varia de acordo com o tamanho do grupo)