Modeling methyl-sensitive transcription factor motifs with an
Por um escritor misterioso
Descrição
Europe PMC is an archive of life sciences journal literature.

Quantitative Analysis of the DNA Methylation Sensitivity of Transcription Factor Complexes - ScienceDirect

Modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet
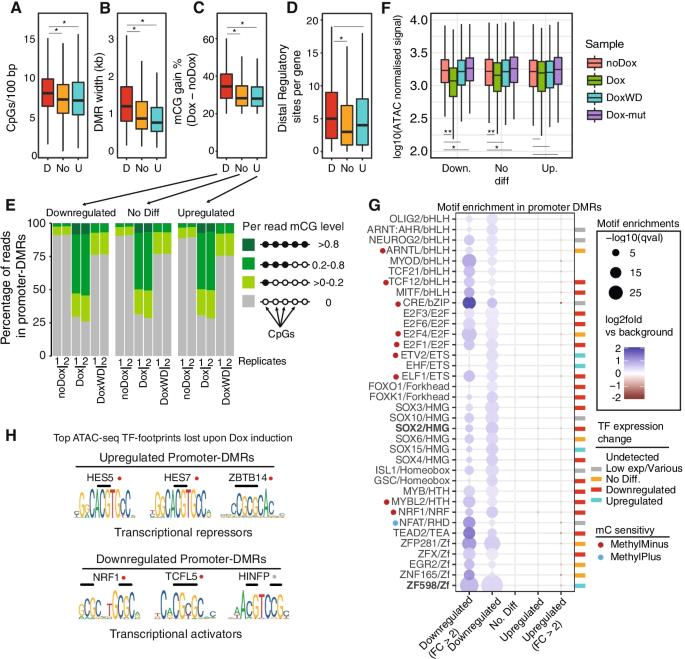
Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability, Genome Biology

Sequence-Specific Structural Features and Solvation Properties of Transcription Factor Binding DNA Motifs: Insights from Molecular Dynamics Simulation

Abnormal methylation in the NDUFA13 gene promoter of breast cancer cells breaks the cooperative DNA recognition by transcription factors, QRB Discovery
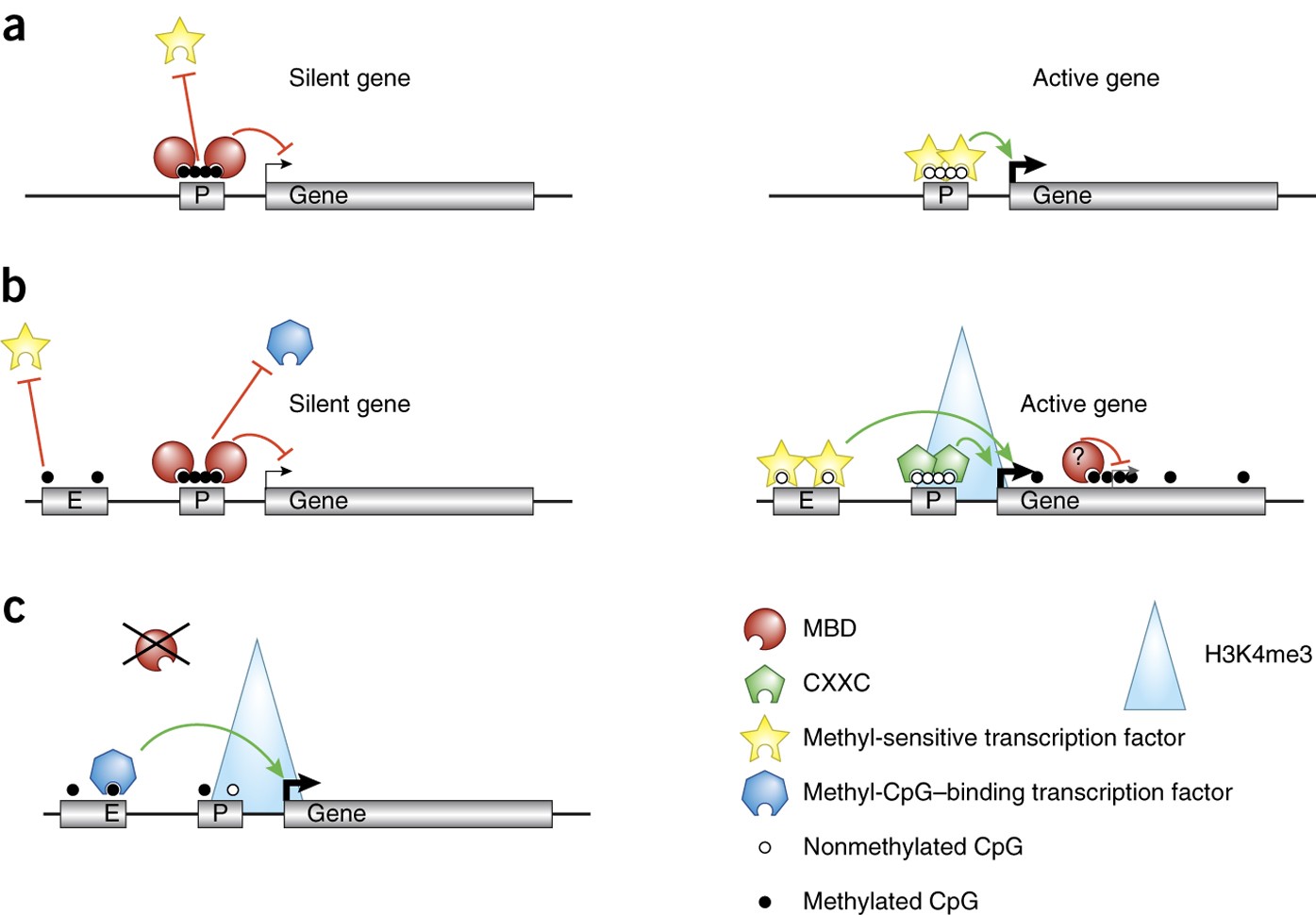
DNA methylation: old dog, new tricks? Nature Structural & Molecular Biology

A simplified model of DNA methylation-mediated gene silencing. (A).

DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contexts
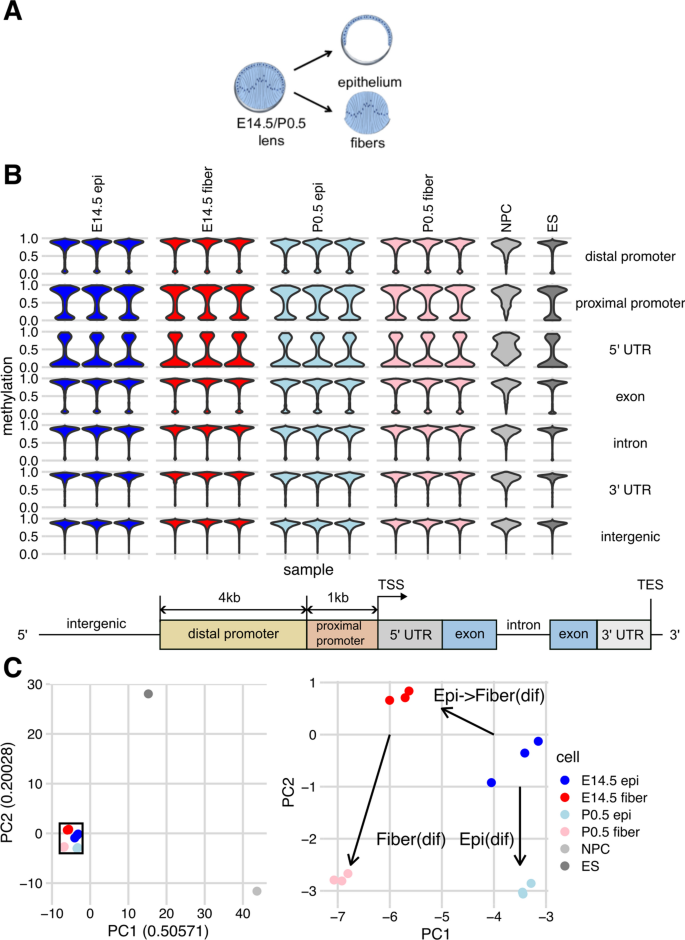
Dynamic changes in whole genome DNA methylation, chromatin and gene expression during mouse lens differentiation, Epigenetics & Chromatin

Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation
Possible scenarios by which TF binding could impact DNA methylation (A)
de
por adulto (o preço varia de acordo com o tamanho do grupo)